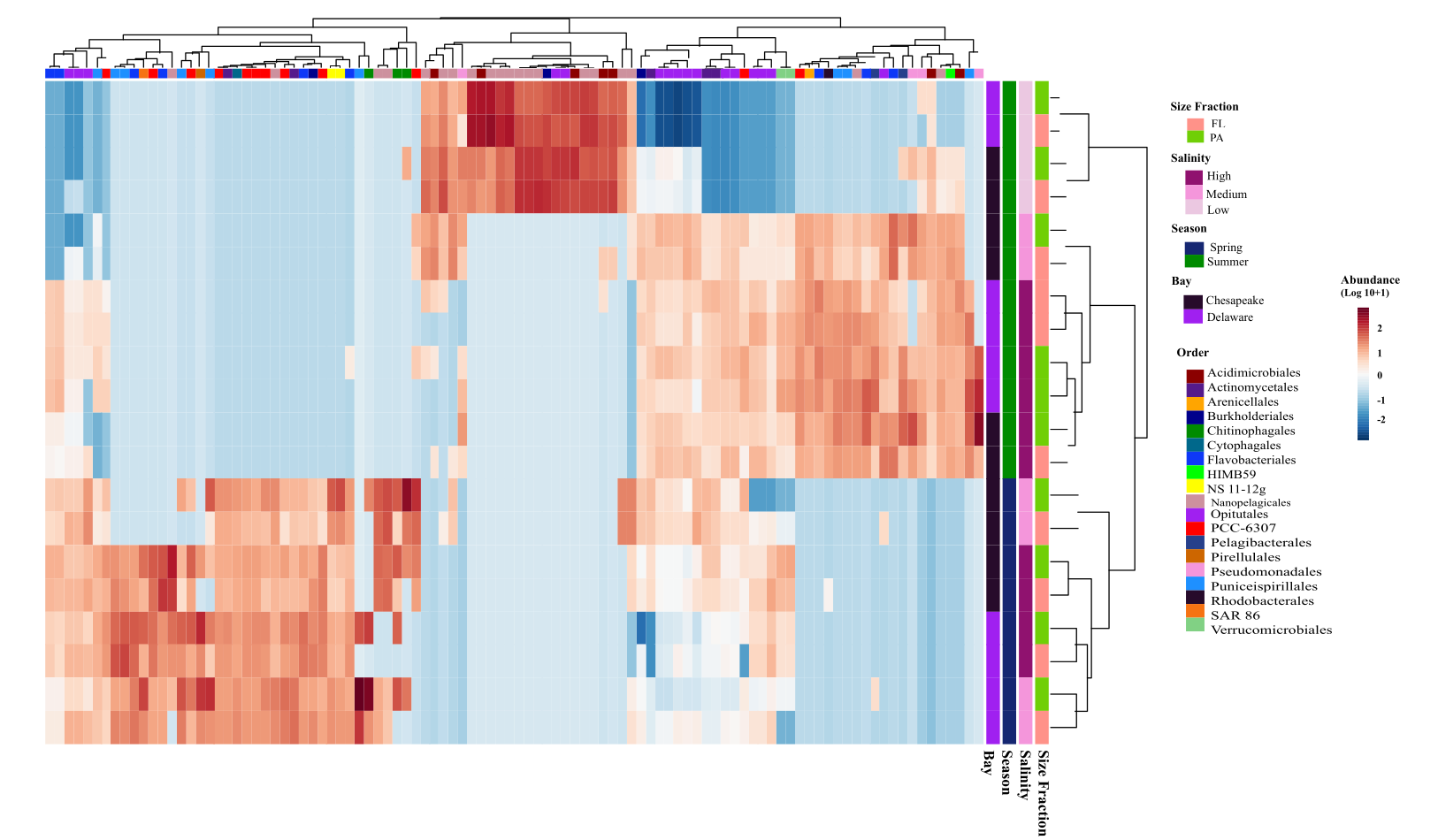
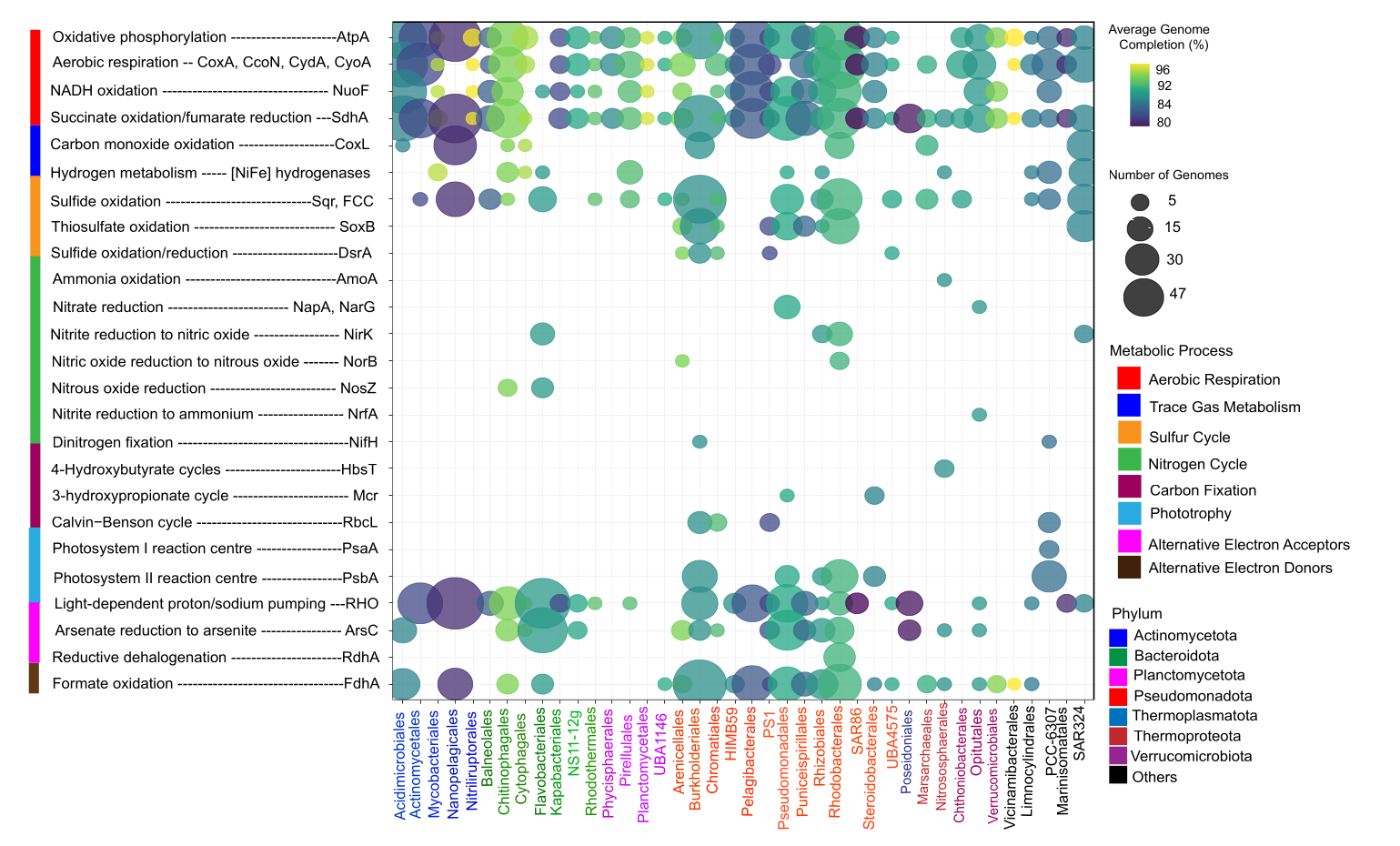
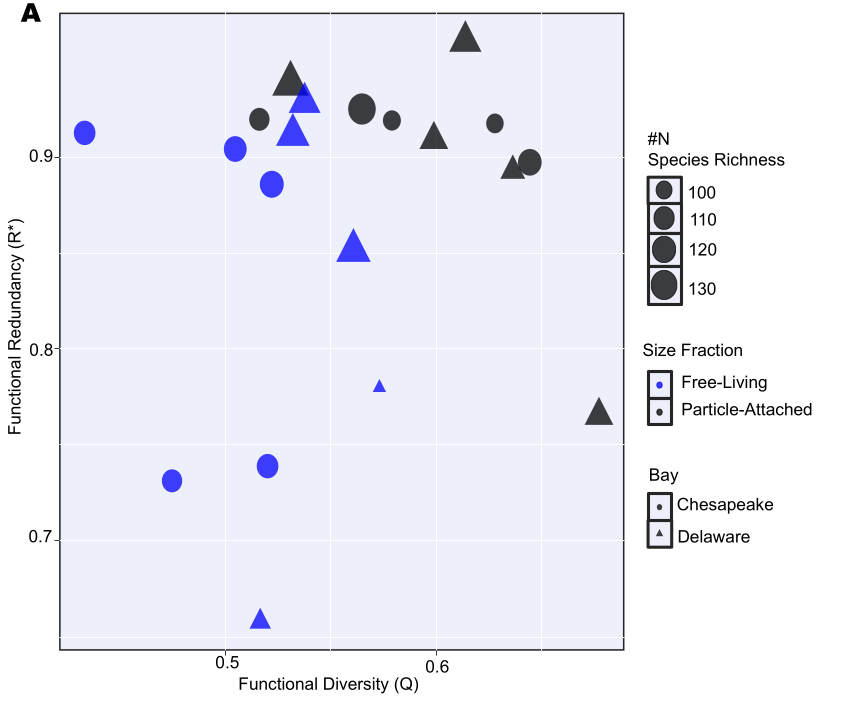
🧠 Background & Motivation
Species and communities respond differently to environmental stress depending on the extent of functional redundancy within the system. In microbial ecosystems, functional redundancy (FRed) can buffer ecosystem processes against disturbance by allowing multiple taxa to perform similar functions. However, the relationship between functional redundancy, metabolic flexibility, and ecosystem resilience remains poorly understood, particularly in dynamic coastal systems.
Estuaries are ideal natural laboratories for studying these relationships. They experience rapid environmental fluctuations driven by seasonality, nutrient inputs, salinity gradients, hypoxia, and anthropogenic stress, often leading to long-term ecosystem degradation. Comparing geographically proximate yet biologically distinct estuaries allows disentangling how community composition and function respond to similar stressors.
In this project, I integrated metagenomics, metatranscriptomics, and genome-resolved analyses across seasonal samples to quantify microbial composition, metabolic potential, activity, and functional redundancy in the Chesapeake and Delaware Bays (USA). This work combines ecological theory with large-scale multi-omics data and statistical modeling to explore how microbial communities maintain ecosystem function under disturbance.
🎯 Research Questions & Objectives
👨🔬 My Role
This project represents my primary postdoctoral research.
🧩 Challenges & Solutions
Challenge 1: Transitioning to fully independent, large-scale bioinformatics without senior technical support
Solution: Systematically evaluated and benchmarked multiple pipelines, optimizing workflows using a subset of samples before scaling analyses to full datasets.
Challenge 2: Lack of established, standardized methods for quantifying microbial functional redundancy
Solution: Developed a customized analytical framework through extensive literature review, interdisciplinary collaboration, and iterative discussions with ecologists and statisticians.
Challenge 3: Interpreting functional redundancy in the absence of directly comparable prior studies
Solution: Integrated environmental metadata, diversity metrics, and metabolic pathway analyses to contextualize FRed patterns within known estuarine gradients and ecological theory.
Challenge 4: Integrating DNA-based potential with RNA-based activity across uneven sequencing depths
Solution: Applied normalization strategies, pathway-level aggregation, and comparative DNA:RNA analyses to robustly link metabolic capacity and expression.
🛠 Methods & Tools
*Data & Sequencing
*Bioinformatics & Statistics
*Tools & Software
*Languages & Workflow
📄 Publications
🎤 Conferences & Talks
Functional redundancy and metabolic flexibility of microbial communities in two Mid-Atlantic bays
Jojy John, Maximiliano Ortiz, Barbara J. Campbell
ASLO Aquatic Sciences Meeting, Charlotte, USA, 2025
Microbial functional redundancy in response to substrate and energy utilization in estuarine ecosystems
Jojy John, Maximiliano Ortiz, Pierre Ramond, Barbara J. Campbell
ISME19, Cape Town, South Africa, 2024
Does the microbiome insure ecosystem function?
Jojy John, Maximiliano Ortiz, Barbara J. Campbell
Clemson University 3rd Postdoctoral Symposium, South Carolina, USA, 2024
🧑🔬 Collaborators / References
Dr. Barbara J. Campbell
Dean’s Distinguished Professor
Department of Biological Sciences, Clemson University
Email: bcampb7@clemson.edu
Dr. Pierre Ramond
Postdoctoral Scientist
Institut de Ciències del Mar (ICM-CSIC)
Email: pierre@icm.csic.es
🖼 Image Gallery


