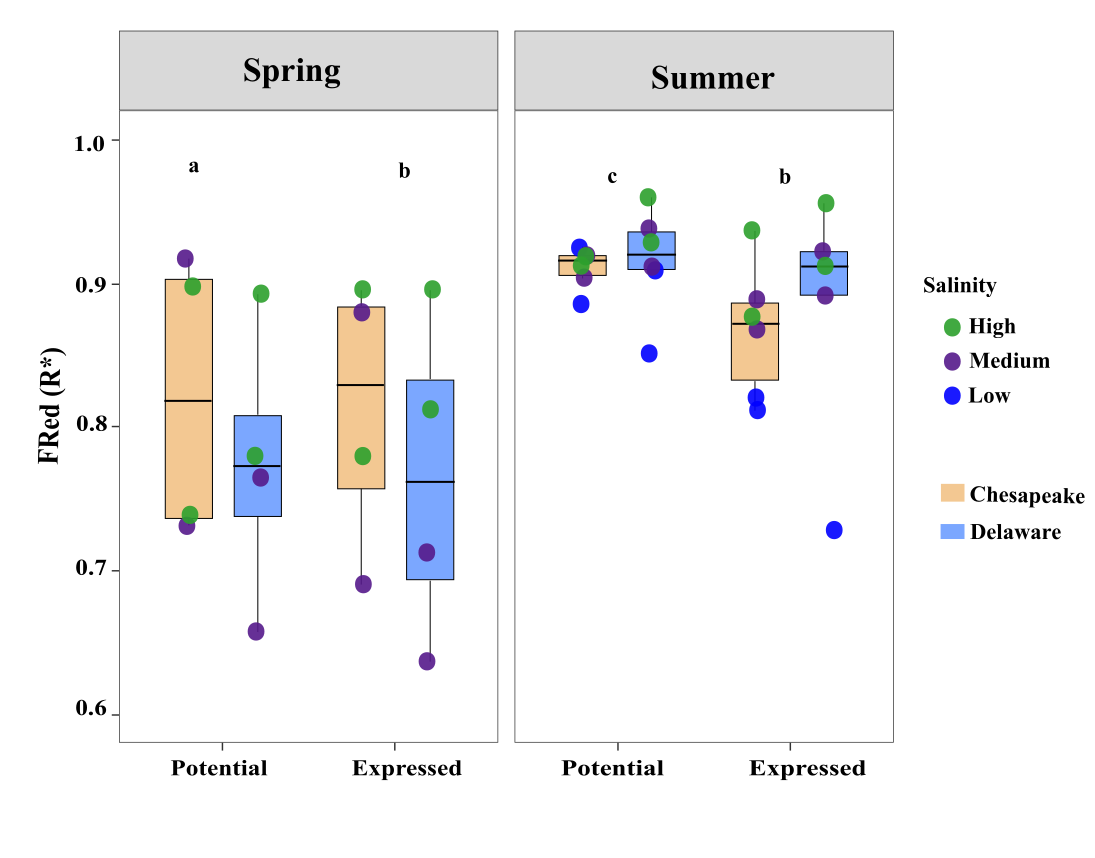
Re-working Figure 5: Visualizing Functional Redundancy (FRed) Across Bays, Seasons, and Salinity
Today I worked on the final revision of one of the main figures (Figure 5) in my manuscript on microbial functional redundancy (FRed).
The goal was to create a clean, publication-quality visualization comparing:
- Potential vs. Expressed FRed (R*)
- Across Chesapeake vs Delaware Bay
- Separated by Spring vs Summer
- Points colored by salinity (Low / Medium / High)
- Boxplots grouped by Bay
- And significance letters (a, b, c) added manually
🔧 Step 1 — Data Cleaning & Long Format Setup
df <- read_excel("pot_Fred_and_exp_Fred.xlsx")
df2 <- df %>%
rename(
Bay = Bays,
Potential = potential,
Expressed = Expressed
) %>%
mutate(Expressed = ifelse(Expressed < 0.25, NA, Expressed)) %>%
pivot_longer(
cols = c("Potential", "Expressed"),
names_to = "Type",
values_to = "FRed"
) %>%
mutate(
Type = factor(Type, levels = c("Potential", "Expressed")),
Season = factor(Season, levels = c("Spring", "Summer")),
Bay = factor(Bay, levels = c("Chesapeake", "Delaware"))
)
🔧 Step 2 — Final Figure (Boxplots + Salinity Colors + Significance Letters)
pd <- position_dodge(width = 0.8)
sig_letters <- data.frame(
Season = factor(c("Spring","Spring","Summer","Summer"),
levels = c("Spring","Summer")),
Type = factor(c("Potential","Expressed","Potential","Expressed"),
levels = c("Potential","Expressed")),
label = c("a","c","b","c"),
y_pos = c(1.02,1.02,1.02,1.02)
)
ggplot(df2, aes(x = Type, y = FRed, fill = Bay)) +
geom_boxplot(
width = 0.55,
color = "black",
outlier.shape = NA,
position = pd
) +
geom_point(
aes(color = Salinity, group = Bay),
shape = 16,
size = 4,
alpha = 0.9,
position = position_jitterdodge(
jitter.width = 0.10,
jitter.height = 0,
dodge.width = 0.8
)
) +
geom_text(
data = sig_letters,
aes(x = Type, y = y_pos, label = label),
inherit.aes = FALSE,
size = 6,
fontface = "bold"
) +
scale_fill_manual(values = c(
"Chesapeake" = "#f3c892",
"Delaware" = "#7ba7ff"
)) +
scale_color_manual(
breaks = c("High", "Medium", "Low"),
values = c(
"High" = "#2ca02c",
"Medium" = "purple4",
"Low" = "blue"
)
) +
facet_wrap(~ Season, nrow = 1) +
coord_cartesian(ylim = c(0.6, 1.05)) +
labs(
x = "",
y = "Functional Redundancy (R*)"
) +
theme_bw(base_size = 16) +
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
strip.text = element_text(size = 18, face = "bold"),
axis.text.x = element_text(size = 14, face = "bold"),
legend.title = element_blank(),
legend.text = element_text(size = 12)
) +
guides(
fill = guide_legend(override.aes = list(shape = NA, color = NA), order = 2),
color = guide_legend(order = 1)
)
🎨 Step 3 — Final Touches in Adobe Illustrator
After saving as an SVG, I edited:
- Font sizes
- Gridlines
- Box spacing
- Alignment
- Exported at 300 dpi
📌 Summary
This workflow produced a clean, publication-ready figure:
- Chesapeake vs Delaware
- Seasonal contrast
- Salinity structure
- Potential vs expressed redundancy
- Added significance letters (a, b, c)