Visualizing Genome Assemblies with Bandage: Building from Source on Linux
🧬 Visualizing Genome Assemblies with Bandage (Built from Source)
Today’s post is all about Bandage — one of the most useful tools for inspecting genome assembly graphs.
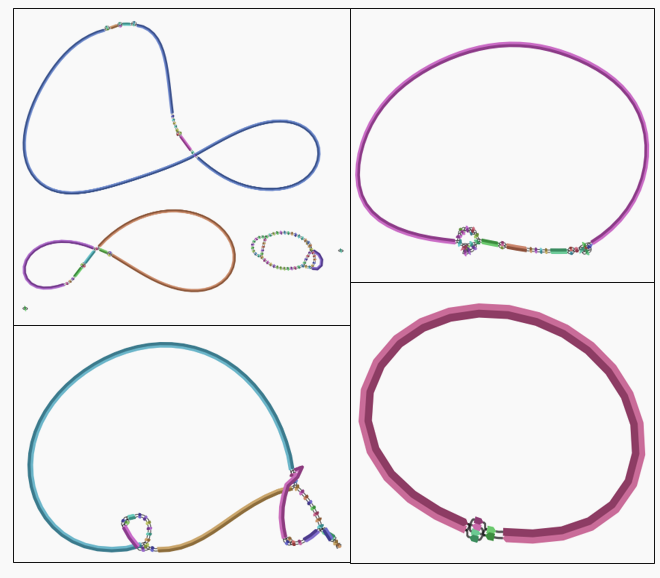
Bandage reads the .gfa output files from assemblers like Flye, SPAdes, and Shovill, allowing you to:
- Visualize contig connections
- Detect circular genomes
- Identify repeats and structural variations
- Inspect gaps, coverage, node lengths
- Export publication-quality images
When I started using hybrid assembly; I also explored their assembly graphs to confirm circularization and check for structural complexity and compare it with individual assemblies from Shovill (Illumina) and Flye (Nanopore).
🔧 Why I Built Bandage from Source
The precompiled Bandage binaries often fail on modern Linux (Ubuntu 24.04), mainly due to:
- missing Qt dependencies
- incompatible system libraries
- version mismatches
So I compiled Bandage manually using Qt 5.15, which is the most stable route for HPC/Linux users.
⚙️ Step 1 — Install Build Tools (Ubuntu)
sudo apt update
sudo apt install -y build-essential git libgl1-mesa-dev libxcb-xinerama0
These provide compilers, OpenGL libraries, and X11 components required for Qt applications.
⚙️ Step 2 — Install Qt SDK (Qt 5.15+ Required)
Bandage requires Qt version 5.15 or later, but Qt 6 is not compatible on all systems.
I installed Qt using the official online installer:
👉 https://www.qt.io/download-qt-installer
During installation:
-
Selected Qt 5.15.x
-
Enabled Desktop GCC 64-bit
-
Installed Qt Creator + core libraries
This step is essential: Bandage will not build without a full Qt environment.
🧰 Step 3 — Open Project in Qt Creator
● Launch Qt Creator
● Open → Bandage.pro from the cloned directory
● Click Configure Project
● Change build mode from Debug → Release
● Click the green ▶ (Run) button
● Qt Creator automatically compiles Bandage and launches the program.
🧬 Step 4 — Viewing Assembly Graphs
Flye and Shovill both generate .gfa files that Bandage can load:
● Flye → assembly_graph.gfa
● Shovill/SPAdes → contigs.gfa
To load a graph:
-
Open Bandage
-
File → Load Graph
-
Select your .gfa file
-
Zoom, rotate, edit labels as needed
🧭 Summary
Today’s work:
✔ Installed Qt + built Bandage from source
✔ Loaded .gfa files from Flye & Shovill
✔ Verified circular genomes
✔ Inspected repeats and graph complexity
✔ Exported clean visualizations for tomorrow’s analysis
Bandage remains an essential tool — especially when you need to see your assemblies rather than just trust metrics like N50.
Stay tuned!
Reference
https://github.com/rrwick/Bandage/wiki